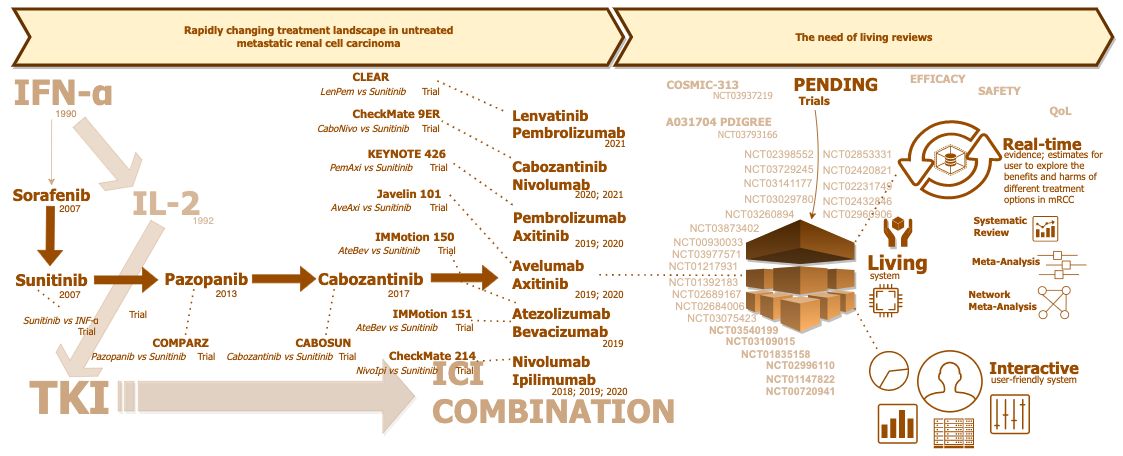
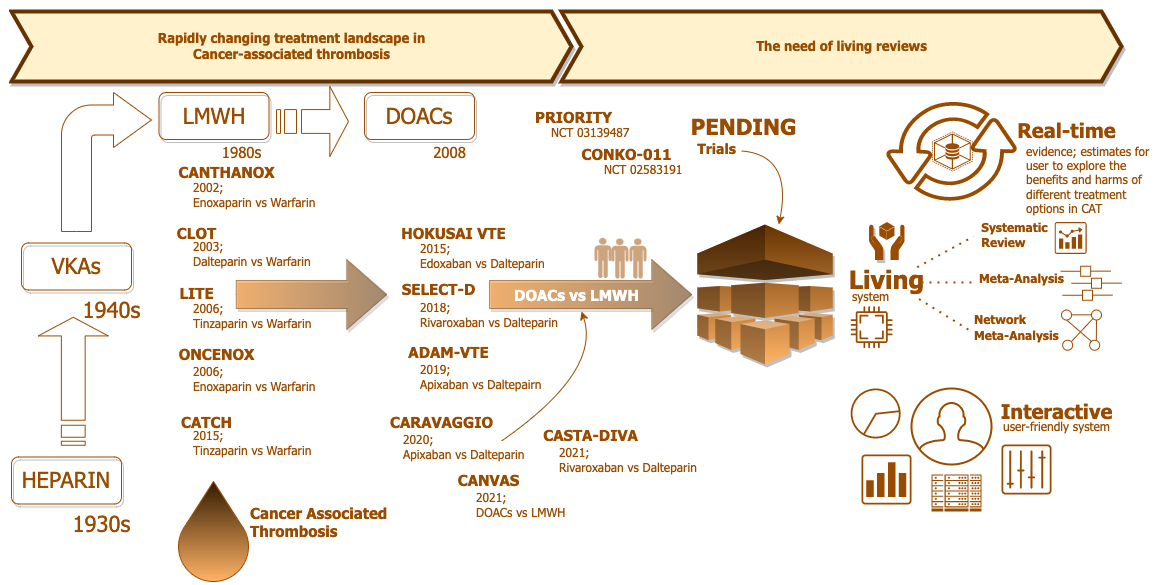
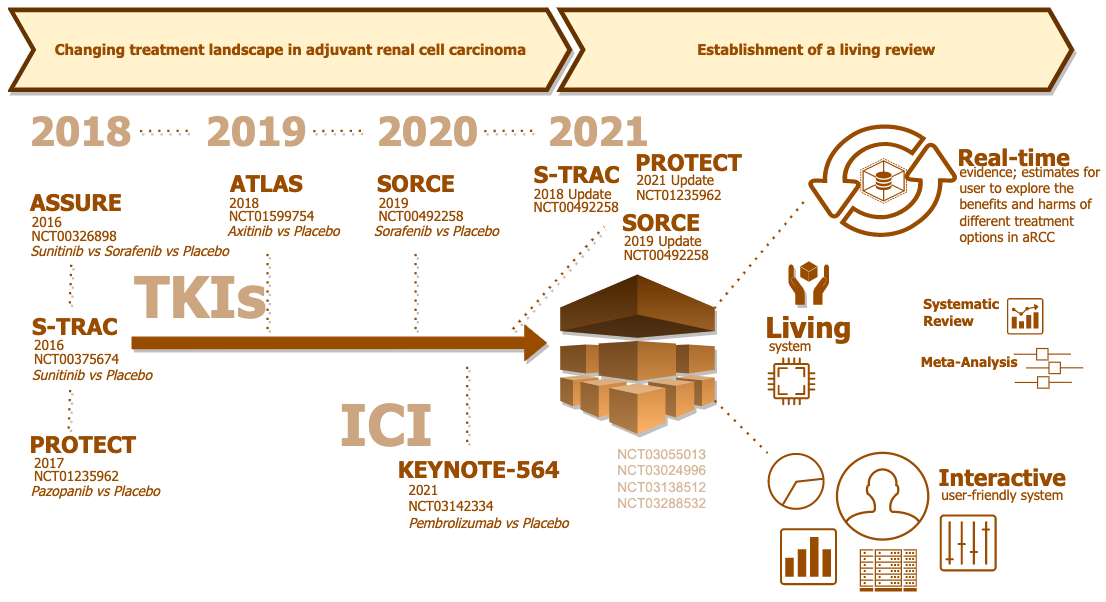
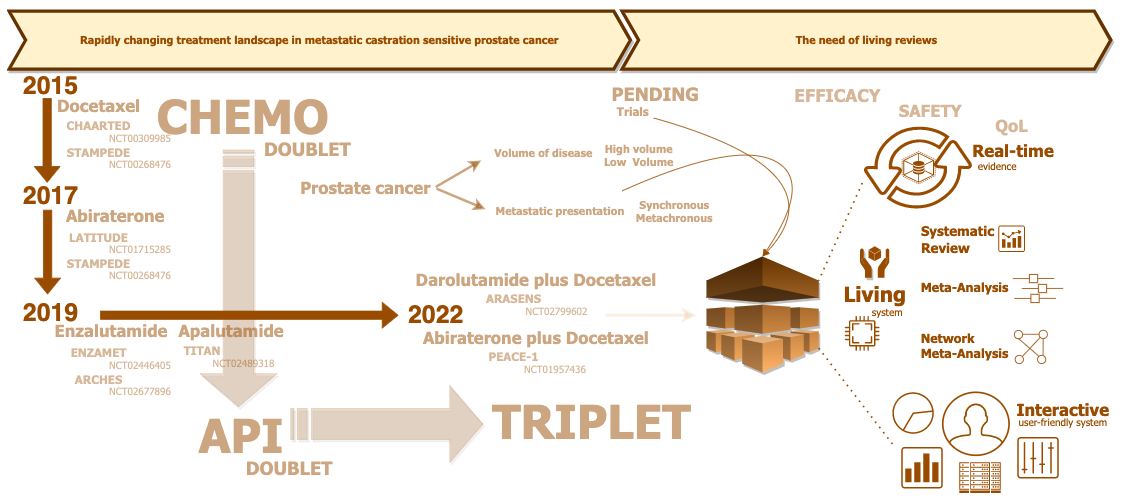
CONCEPT
Systematic reviews (SR) and meta-analyses (MA) are tools to synthesize evidence and provide decision makers with estimates of effect that are more precise than those provided by individual studies. SRMAs inform decision makers about the certainty in these estimates to allow tradeoffs and inform shared decision making. SRMAs also support clinical practice guidelines (CPGs) and Institute of Medicine (IOM) and Guideline International Network (G-I-N) require an updated systematic review as one of the criteria for trustworthy clinical practice guidelines (CPGs).
There are two major limitations in the current approach to evidence synthesis. The process of creating SRMAs is cumbersome and slow and the presentation of results using conventional static tables and figures limits the depth of information that can be informative to clinicians and evidence users. Consequence is an epidemic of redundant, conflicted systematic reviews and meta-analyses (SRMAs). In areas with rapidly moving evidence, many SRMAs are outdated as soon as they are published. Usually, there is little incentive for original team to undertake the laborious updating process and hence a completely new time will try to create (update) an SRMA from the scratch. Often, there are just too many of redundant, overlapping systematic reviews.
Some examples are atrocious, such as patent foramen ovale (PFO) closure management example; where there are dozens meta-analyses synthesizing the information from four published randomized clinical trials. This duplication of effort is not only wasteful but often results in conflicted findings due to subtle differences in design or analysis strategy.
Living Systematic Reviews—which are continually updated, incorporating relevant new evidence as it becomes available—has been suggested as a solution to address the challenge of synthesizing evidence in fields with rapidly moving evidence. Leading journals such as Annals and BMJ have welcomed this approach where authors commit to frequent updates on accepted systematic reviews. However, without a framework supported by advanced programming and Artificial intelligence, the approach of living systematic reviews is not “truly” living and merely represents an effort of undertaking a conventional systematic review with a commitment to frequent dates. While the latter is a step in the right direction but only reflects a part of the solution. Finally, SRMAs often have multiple tables, figures and analyses hidden in the supplemental materials with no user-friendly access. This alarming increase in wasteful efforts for SRMAs of minimal value warrants reconsideration of methods, production, and reporting of SRMAs. Hence, we propose a Living Interactive Evidence Synthesis (LIvE) framework as an approach to create Living interactive Systematic Reviews (LISRs).